Comparison of Python Distributions on Apocrita¶
When it comes to picking a distribution, Python programmers are spoilt for choice. We're going to compare two of the most popular (CPython and Anaconda) and one that promises big performance improvements with relatively little hassle (Intel Distribution for Python).
If you haven't come across it before, Intel Distribution for Python is a
distribution from Intel with a data-science focus. It is similar to Anaconda
in that it includes popular scientific libraries and uses conda for package
management. Intel have a conda channel, on
which they host their own versions of several of these common libraries.
On the face of it, we might expect our three distributions to perform identically, since they all use the same CPython interpreter, as you can see with:
>>> import platform
>>> platform.python_implementation()
'CPython'
For programs written in pure Python, performance might indeed be identical. However, in the kind of computing done on the HPC cluster here at QMUL, a large proportion of the time is spent in calls to numerical libraries such as numpy, scipy and scikit-learn. Since different Python distributions come with different versions of these libraries, we may expect to find that some are quicker than others.
The main difference between, for example, the CPython Numpy and Anaconda Numpy packages will be the underlying maths libraries. Just as scikit-learn builds on scipy and scipy in turn builds upon numpy, numpy builds upon the low-level routines of other packages such as BLAS.
BLAS is not a library per se but a specification for other libraries to follow. There are many implementations of BLAS such as openBLAS, ATLAS and Intel's MKL. To some extent, then, our results are going to be indicative of the performance of the underlying low-level maths libraries more that anything inherent in the different Python distributions. However, it is useful to know which distributions come with the most optimised libraries to inform our choices.
Methods¶
On Apocrita, QMUL's HPC cluster, we set up three Python virtual environments; one each for CPython, Anaconda and Intel Python. QMUL readers can see the environment files in our rse-python-benchmarks repository. Into these environments, we installed Intel's ibench benchmarking suite and its dependencies. You can clone it from Intel's ibench repository or from our fork of it. Our fork adjusts the size of some of the tests so that they don't use too much memory and removes some deprecated parameters from function calls but the results should be the same either way.
We ran the tests as HPC jobs, with separate jobs for one and four cores to see whether some distributions scale better than others. Within a job, the tests ran sequentially (CPython, followed by Anaconda and then Intel Python).
Results¶
Figure 1, below, is representative of our first attempt. Notice the log scale, which means that one horizontal line higher equates to taking 10 times longer to complete the task.
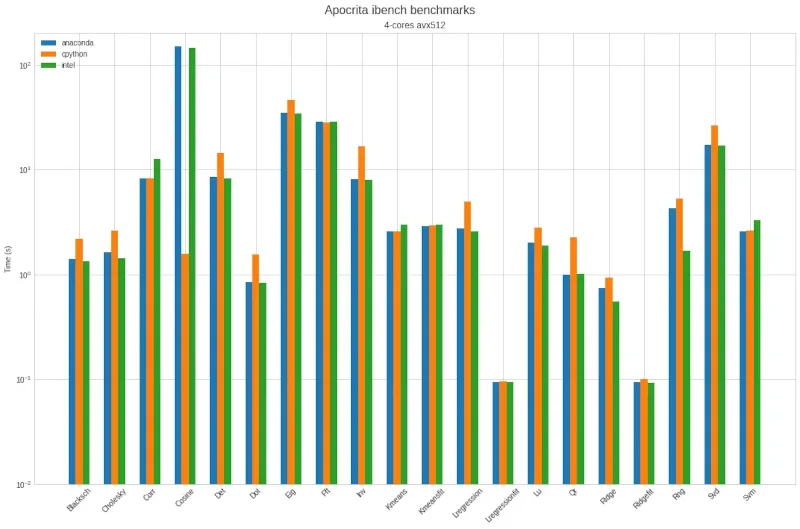 Figure 1: First benchmark results on Apocrita showing significantly different behaviours on the Cosine test.
Figure 1: First benchmark results on Apocrita showing significantly different behaviours on the Cosine test.
The two things that stand out are:
- Intel Python and Anaconda are closely matched and generally a bit quicker than CPython
- Intel Python and Anaconda are considerably slower on the Cosine test
We investigated the surprising slowness of Anaconda and Intel Python and found
that some of our Intel CPUs support the AVX-512 instruction set. Either a bug
or an unfortunate combination of array sizes, environment variables and thread
numbers means that matrix multiplication with four threads on an AVX-512
enabled CPU grinds to a near halt. The good news is that turning off the
instruction set with MKL_ENABLE_INSTRUCTIONS=AVX2 or changing the number of
threads +/-1 with either the OMP_NUM_THREADS or MKL_NUM_THREADS environment
variables will work around the issue.
While we were investigating the case and solution of the slowdown, we found other environment variables that can be used to improve performance. Specifically, the version of scikit-learn you get with Intel Python gives an easily-missed message on installation
INSTALLED PACKAGE OF SCIKIT-LEARN CAN BE ACCELERATED USING DAAL4PY.
PLEASE SET 'USE_DAAL4PY_SKLEARN' ENVIRONMENT VARIABLE TO 'YES' TO ENABLE THE ACCELERATION.
On our next run, seen in Figure 2 below, we set OMP_NUM_THREADS=5 to avoid the
AVX-512 issue and tried Intel Python both with and without setting
USE_DAAL4PY_SKLEARN=YES.
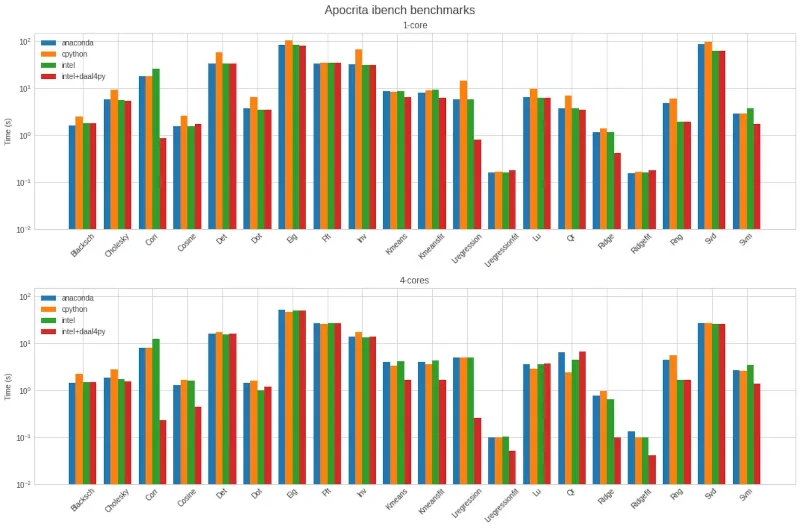 Figure 2: Second benchmark results showing greater consistency on all tests.
Figure 2: Second benchmark results showing greater consistency on all tests.
Here we see that:
- Setting
USE_DAAL4PY_SKLEARN=YESgives a large performance improvement on several tests, particularly Corr (sklearn pairwise distances using the correlation metric) and Lregression (fitting a linear regression model) - When not using
USE_DAAL4PY_SKLEARN=YES, the performance difference between CPython, Anaconda and Intel Python is even smaller for four cores than for one - Anaconda and Intel Python performance is now on par with CPython for the Cosine test, showing that the AVX-512 issue has been circumvented
The similarity in performance between Anaconda and Intel Python (and their slight improvement over CPython) is at least partly explained by them both including versions of Numpy linked to Intel's MKL libraries (whereas CPython's Numpy uses OpenBLAS). You can get some information from Numpy about the libraries it was built with by starting Python and typing
import numpy
numpy.show_config()
which, if you're using CPython, will give something like
...
blas_mkl_info:
NOT AVAILABLE
blis_info:
NOT AVAILABLE
openblas_info:
libraries = ['openblas', 'openblas']
library_dirs = ['/usr/local/lib']
language = c
define_macros = [('HAVE_CBLAS', None)]
...
and, if you are using Anaconda or Intel Python, something like
...
blas_mkl_info:
libraries = ['mkl_rt', 'pthread']
define_macros = [('SCIPY_MKL_H', None), ('HAVE_CBLAS', None)]
blas_opt_info:
libraries = ['mkl_rt', 'pthread']
define_macros = [('SCIPY_MKL_H', None), ('HAVE_CBLAS', None)]
...
Conclusions¶
The Benefits of Benchmarking¶
Benchmarks aren't always predictive of real-world performance, where other users of the system, network/disk performance and so on all conspire to confound expectations. However, they have proven useful because:
- Benchmarks help to uncover bugs, hardware issues and installation errors
- By comparing Anaconda, Intel Python and CPython, we could get a feel for the relative performance of the three, without worrying about absolute numbers
Recommendations¶
We had hopes that Intel Python would provide an appreciable speed up for a very small support cost. However, we did not find a significant improvement for most tasks. While not difficult, Intel Python is neither as easy to install nor to configure as Anaconda or CPython. The benchmarks we ran weren't comprehensive enough to conclude that Intel Python wouldn't make a noticeable difference to your project. However, if you are considering using it, we would recommend doing your own benchmarks (with representative tests) first.
Jan Hynek drew similar conclusions when he ran ibench and other benchmarks, which you can read about here.

